This function calculates evolutionary distinctiveness according to the fair-proportion index for each species.
References
Isaac, N. J., Turvey, S. T., Collen, B., Waterman, C. and Baillie, J. E. (2007). Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE 2, e296.
Examples
# \donttest{
library(phyloraster)
tree <- ape::read.tree(system.file("extdata", "tree.nex",
package="phyloraster"))
plot(tree)
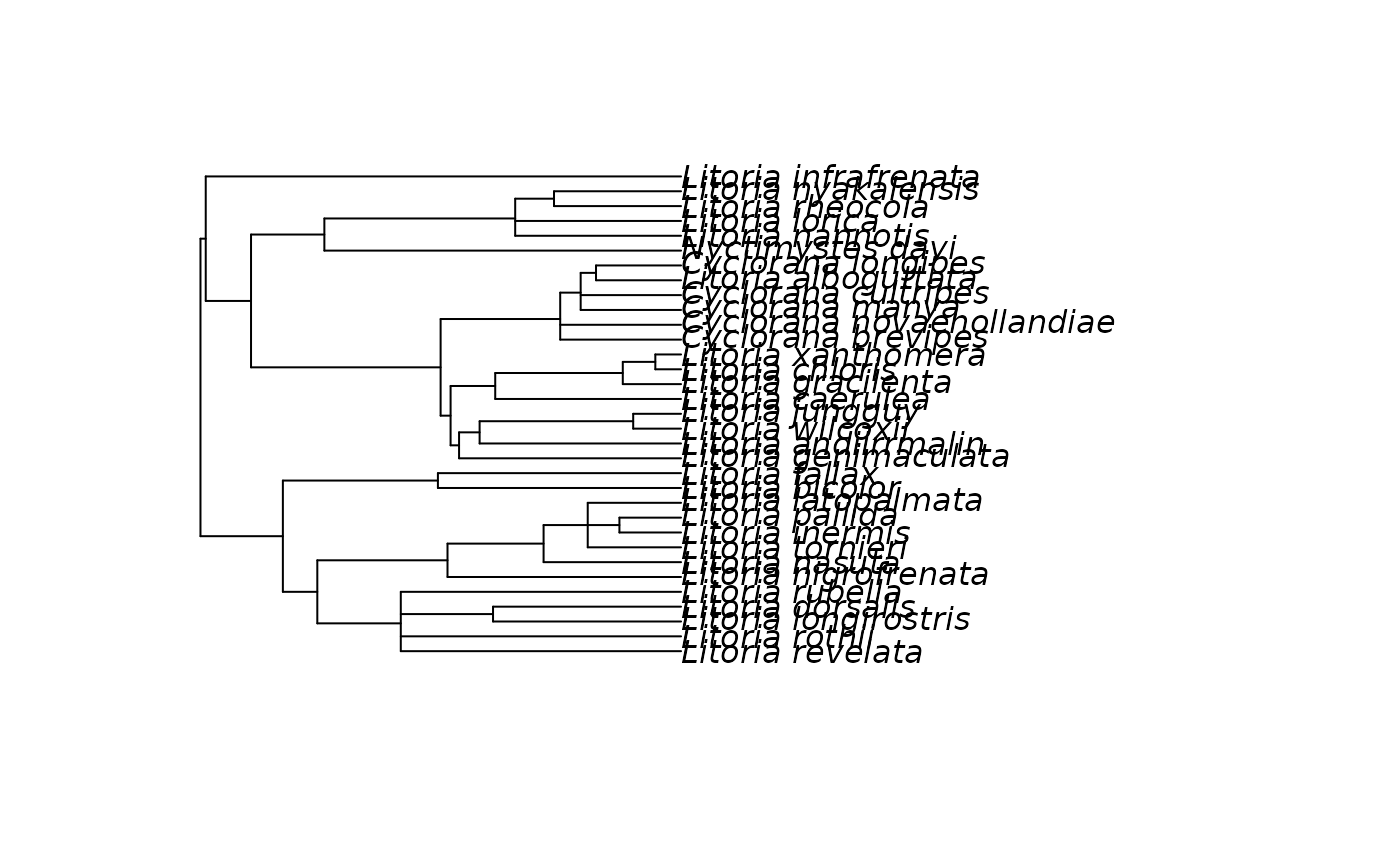 ed <- species.ed(tree)
ed
#> ED
#> Litoria_revelata 0.6440047
#> Litoria_rothii 0.6440037
#> Litoria_longirostris 0.5470682
#> Litoria_dorsalis 0.5470682
#> Litoria_rubella 0.6440037
#> Litoria_nigrofrenata 0.5563398
#> Litoria_nasuta 0.3947966
#> Litoria_tornieri 0.3252204
#> Litoria_inermis 0.2919079
#> Litoria_pallida 0.2919079
#> Litoria_latopalmata 0.3252204
#> Litoria_bicolor 0.6872423
#> Litoria_fallax 0.6872423
#> Litoria_genimaculata 0.5076379
#> Litoria_andiirrmalin 0.4788422
#> Litoria_wilcoxii 0.3174512
#> Litoria_jungguy 0.3174512
#> Litoria_caerulea 0.4506342
#> Litoria_gracilenta 0.2719152
#> Litoria_chloris 0.2377227
#> Litoria_xanthomera 0.2377227
#> Cyclorana_brevipes 0.3298920
#> Cyclorana_novaehollandiae 0.3298920
#> Cyclorana_manya 0.2976395
#> Cyclorana_cultripes 0.2976405
#> Litoria_alboguttata 0.2815130
#> Cyclorana_longipes 0.2815130
#> Nyctimystes_dayi 0.7860100
#> Litoria_nannotis 0.4852217
#> Litoria_lorica 0.4852217
#> Litoria_rheocola 0.4444367
#> Litoria_nyakalensis 0.4444367
#> Litoria_infrafrenata 0.9995006
# }
ed <- species.ed(tree)
ed
#> ED
#> Litoria_revelata 0.6440047
#> Litoria_rothii 0.6440037
#> Litoria_longirostris 0.5470682
#> Litoria_dorsalis 0.5470682
#> Litoria_rubella 0.6440037
#> Litoria_nigrofrenata 0.5563398
#> Litoria_nasuta 0.3947966
#> Litoria_tornieri 0.3252204
#> Litoria_inermis 0.2919079
#> Litoria_pallida 0.2919079
#> Litoria_latopalmata 0.3252204
#> Litoria_bicolor 0.6872423
#> Litoria_fallax 0.6872423
#> Litoria_genimaculata 0.5076379
#> Litoria_andiirrmalin 0.4788422
#> Litoria_wilcoxii 0.3174512
#> Litoria_jungguy 0.3174512
#> Litoria_caerulea 0.4506342
#> Litoria_gracilenta 0.2719152
#> Litoria_chloris 0.2377227
#> Litoria_xanthomera 0.2377227
#> Cyclorana_brevipes 0.3298920
#> Cyclorana_novaehollandiae 0.3298920
#> Cyclorana_manya 0.2976395
#> Cyclorana_cultripes 0.2976405
#> Litoria_alboguttata 0.2815130
#> Cyclorana_longipes 0.2815130
#> Nyctimystes_dayi 0.7860100
#> Litoria_nannotis 0.4852217
#> Litoria_lorica 0.4852217
#> Litoria_rheocola 0.4444367
#> Litoria_nyakalensis 0.4444367
#> Litoria_infrafrenata 0.9995006
# }
